Detecting Potential Cardiovascular Inflammation through Peak Analysis of SERS Data
by Professor Mark Roantree, Insight Funded Investigator, DCU and Insight members Dinh Viet Cuong, Damhan Richardson and Nirod Sarangi
Exosomes are tiny cell-derived vesicles emitted by all cells. They are surrounded by a membrane much like the membrane from their originating cell and carry a diverse cargo of macromolecules including proteins, lipids, mRNAs and microRNAs. As their contents reflect their origin, exosomes can provide insight into the metabolic status and health of the cells they are derived from. Therefore, they are emerging as important diagnostic tools, accessible through liquid biopsy.
In this project, cardiac endothelial cells, the cells that line the blood vessels of the heart, were grown under normal, hypo and hyperglycaemic conditions, the latter intended to model endothelial dysfunction, typical of cardiovascular inflammation. The composition of exosomes emitted by these cells is then analysed using a technique known as surface enhanced Raman spectroscopy (SERS). The spectra are highly complex as they reflect contributions from all constituents of the exosome but provide a“fingerprint” that reflects the originating cell’s health. The classification of these exosomes is a good task for the right machine learning function. As part of the feature extraction process, peak analysis is used to generate profiles for each of the three cells types (hypoglycaemic, hyperglycaemic, and healthy) and is used for learning and validation during the creation of the machine learning model.
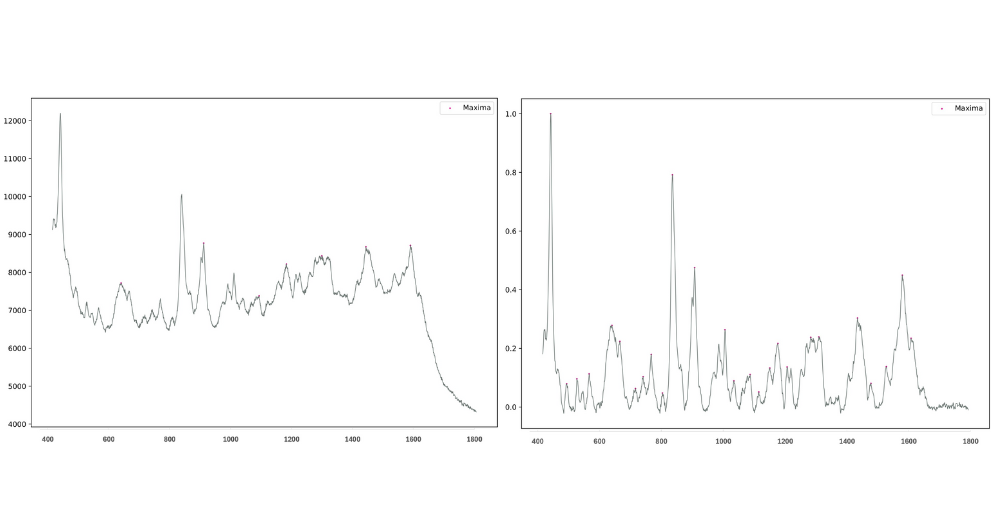
The first figure above represents the Raw SERS spectrum of one sample of hypoglycaemic exosome. The second figure represents the same with pre-processing i.e. scaling, background removal, and smoothing.
During peak analysis, a “peak” profile depends on the settings of: minimum prominence, minimum width, and minimum distance between peaks. Defining a search space of these parameters allows for the generation of several hundred unique feature sets, all of which are fed into the machine learning functions.
This collaborative research effort involves members from Prof. Mark Roantree’s team in the School of Computing, Prof. Tia Keyes’ team in the School of Chemistry, and Prof Paul Cahill in the School of Biotechnology, all at Dublin City University. Team members include Dinh Viet Cuong, Damhan Richardson and Nirod Sarangi from the Insight SFI Reseach Centre for Data Analytics and PhD student Thomas Keogh from the SFI Centre for Research Training in Artificial Intelligence.